在临床试验中,通常使用SAS来完成统计分析和生成图表,但我们不应该只局限于一种编程方法,而且这个所用的编程语言SAS并不是开源的。毫无疑问SAS能完成的事情,R和Python同样能做;但有些R和Python能做的,SAS却很难完成,我想这就是开源和不开源的区别。
之前可能只有在药厂内部的一些需求会使用R或者Python,但现在随着大药厂开始陆陆续续尝试使用R生成的结果来提交给监管机构,以及伴随着递交数据的改变(如转变成json格式),后续R或者Python在临床统计分析中应该会有一个完整的分析-呈现-递交的工作流。
以下是我对于R包rtables的初步尝试,比如我们想生成一张在肿瘤试验中常见的生存分析的表格,如下所示(非递交用途)。
但不得不说假如真要使用rtables包来生成临床分析表格,最好还是搭配tern包来使用;前者主要是生成表格,后者则是用来分析数据,而且是无缝连接。
假如不想使用tern包的话,就得用另外一种使用方式,即写自定义函数来分析,然后在rtables的函数里调用,但这样的话就不如tern + rtables这种方式来得便捷。
步入正题,首先我先加载whas500数据集用于后续的生存分析(Kaplan-Meier),主要用到其中的3个变量
- AFB: Atrial Fibrillation (0 = No, 1 = Yes)
- LENFOL: Total Length of Follow-up (Days between Date of Last Follow-up and Hospital Admission Date)
- FSTAT: Vital Status at Last Follow-up (0 = Alive 1 = Dead)
并对分组变量AFB做一些处理,拟合KM模型,其中置信区间的方法采用log-log转化
library(survival)
library(tidyverse)
library(rtables)
data("whas500", package = "stabiot")
dat <- whas500 %>%
dplyr::mutate(
AFB = case_when(
AFB == 1 ~ "Yes",
TRUE ~ "No"
),
AFB = factor(AFB, levels = c("Yes", "No"))
)
km_fit <- survfit(
data = dat,
formula = Surv(LENFOL, FSTAT) ~ AFB,
conf.int = 0.95,
conf.type = "log-log"
)接着分析中位、25th和75th的生存时间以及log-rank检验,并对分析结果做一些调整使得其能更加容易匹配在rtables语法(即避免在rtables自定义函数中调整数据格式)
# median survival time
surv_med <- summary(km_fit)$table
# quantile survival time
surv_quant <- quantile(km_fit, probs = c(0.25, 0.75)) %>%
purrr::map(\(df) as.data.frame(df) %>%
rownames_to_column(var = "group")) %>%
purrr::list_rbind() %>%
mutate(
stat = rep(c("est", "lower", "upper"), each = 2)
) %>%
pivot_longer(cols = c("25", "75"), names_to = "quantile") %>%
pivot_wider(
names_from = c("stat", "quantile"), names_glue = "Q{quantile}_{stat}",
values_from = "value"
) %>%
column_to_rownames(var = "group")
# test survival curves
surv_pval <- survminer::surv_pvalue(km_fit, method = "log-rank")然后分析在12、36和60月时的生存率以及两组间率的比较,后者主要是通过Z检验来计算CI和P值。
# survival rate at 12/36/60 months
tp_cols <- c("time", "n.risk", "n.event", "n.censor", "surv", "std.err", "lower", "upper")
surv_rate <- summary(km_fit, times = c(12, 36, 60), extend = TRUE)[tp_cols] %>%
as.data.frame() %>%
split(~time) %>%
purrr::map(\(df) magrittr::set_rownames(df, c("AFB=Yes", "AFB=No")))
# difference of survival rate by time-point
surv_rate_diff <- surv_rate %>%
purrr::map(function(x){
tibble::tibble(
time = unique(x$time),
surv.diff = diff(x$surv),
std.err = sqrt(sum(x$std.err^2)),
lower = surv.diff - stats::qnorm(1 - 0.05 / 2) * std.err,
upper = surv.diff + stats::qnorm(1 - 0.05 / 2) * std.err,
pval = if (is.na(std.err)) {
NA
} else {
2 * (1 - stats::pnorm(abs(surv.diff) / std.err))
}
)
})以上是完成了分析的步骤,其实这些常规的分析步骤都已经包括在tern包里了。
接下来则是写自定义的函数,用于在rtables中的analyze()函数。
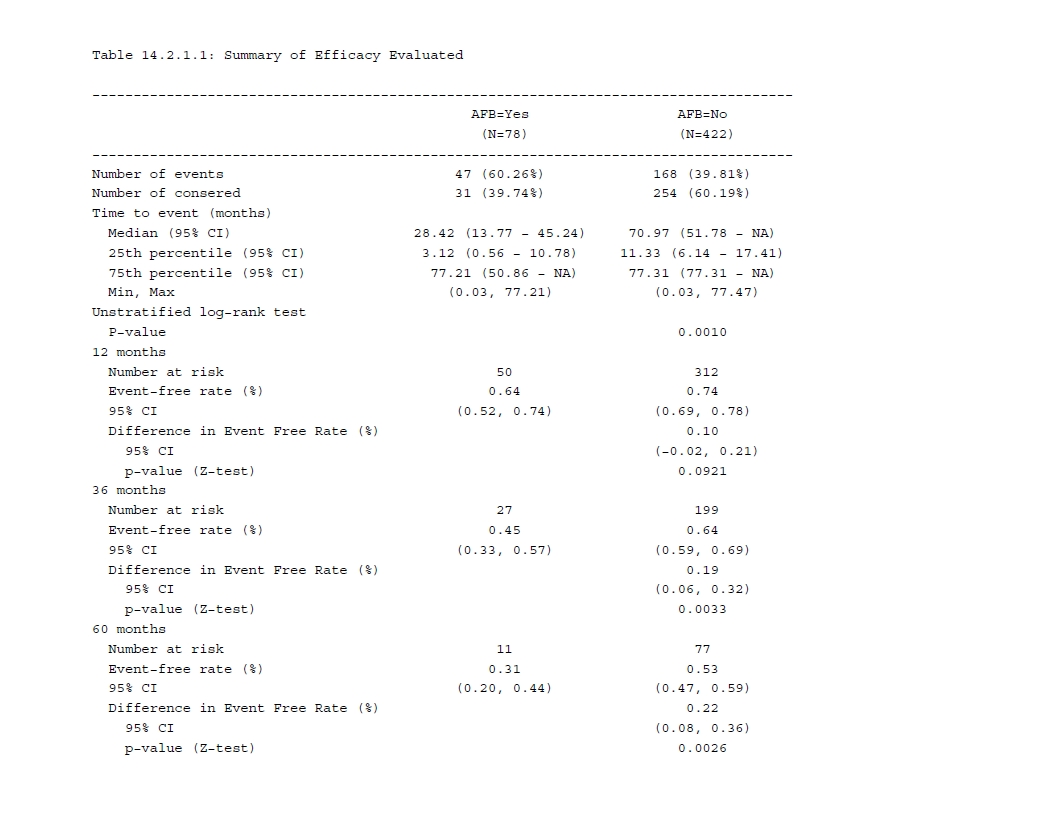
首先汇总两组的事件数和删失数,则需要先计算输入数据集中FSTAT变量的0/1分类数目,然后除以每组人数.N_col来得到百分比。in_rows()代表多行分析,即每个输入代表一行结果,format参数来定义输出格式
a_count_subjd <- function(df, .N_col) {
in_rows(
"Number of events" = rcell(
sum(df$FSTAT == 1) * c(1, 1 / .N_col), format = "xx (xx.xx%)"
),
"Number of consered" = rcell(
sum(df$FSTAT == 0) * c(1, 1 / .N_col), format = "xx (xx.xx%)"
)
)
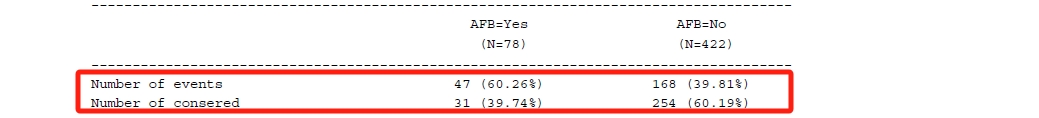
}接着汇总各个分位数下的生存时间估计及其CI,以及生存时间的最大最小值。在这个自定义函数中,我使用了额外参数(med_tb和quant_tb),分别对应中位数和25th/75th分位数的数据集,其中ind变量可用于从上述数据集中找到对应组别的那行结果
a_surv_time_func <- function(df, .var, med_tb, quant_tb) {
ind <- grep(df[[.var]][1], row.names(med_tb), fixed = TRUE)
med_time = list(med_tb[ind, c("median", "0.95LCL", "0.95UCL")])
quantile_time = lapply(c("25", "75"), function(x) {
unlist(c(quant_tb[ind, grep(paste0("Q", x), names(quant_tb))]))
})
range_time = list(range(df[["LENFOL"]]))
in_rows(
.list = c(med_time, quantile_time, range_time),
.names = c(
"Median (95% CI)",
"25th percentile (95% CI)",
"75th percentile (95% CI)",
"Min, Max"
),
.formats = c(
"xx.xx (xx.xx - xx.xx)",
"xx.xx (xx.xx - xx.xx)",
"xx.xx (xx.xx - xx.xx)",
"(xx.xx, xx.xx)"
)
)
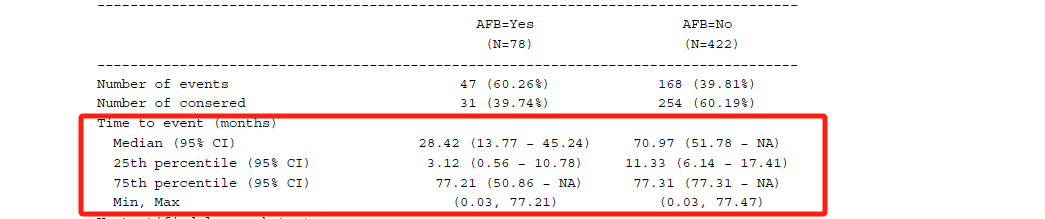
}然后汇总两组的log-rank检验的P值,pval_tb对应其数据集
a_surv_pval_func <- function(df, .var, .in_ref_col, pval_tb) {
in_rows(
"P-value" = non_ref_rcell(
pval_tb[["pval"]],
.in_ref_col,
format = "x.xxxx | (<0.0001)"
)
)
}最后汇总12、36和60月时的生存率以及两组间率的比较,rate_tb和rate_diff_tb分别对应各个time point的生存率和组间率差的数据集。其中non_ref_rcell()可用于reference group需要为空的情况,indent_mod参数则可以调整缩进尺度(默认是0,即不缩进)
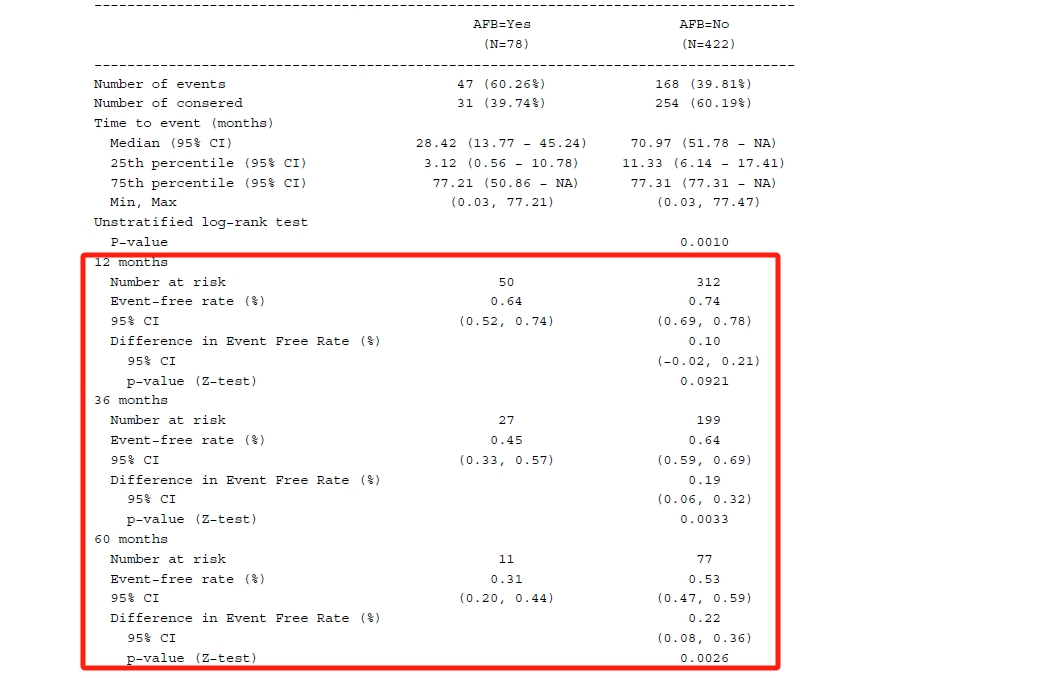
a_surv_rate_func <- function(df, .var, .in_ref_col, rate_tb, rate_diff_tb) {
ind <- grep(df[[.var]][1], row.names(rate_tb), fixed = TRUE)
in_rows(
rcell(rate_tb[ind, "n.risk", drop = TRUE], format = "xx"),
rcell(rate_tb[ind, "surv", drop = TRUE], format = "xx.xx"),
rcell(unlist(rate_tb[ind, c("lower", "upper"), drop = TRUE]), format = "(xx.xx, xx.xx)"),
non_ref_rcell(
rate_diff_tb[, "surv.diff", drop = TRUE],
.in_ref_col,
format = "xx.xx"
),
non_ref_rcell(
unlist(rate_diff_tb[, c("lower", "upper"), drop = TRUE]),
.in_ref_col,
format = "(xx.xx, xx.xx)",
indent_mod = 1L
),
non_ref_rcell(
rate_diff_tb[, "pval", drop = TRUE],
.in_ref_col,
format = "x.xxxx | (<0.0001)",
indent_mod = 1L
),
.names = c(
"Number at risk",
"Event-free rate (%)", "95% CI",
"Difference in Event Free Rate (%)", "95% CI",
"p-value (Z-test)"
)
)
}完成上述各个自定义函数后,接着是则进入rtables的layout部分,由于我们的这次的表格比较简单,所以所用的函数不复杂。basic_table()完成基本表格元素的设定,如title和footnote等;split_cols_by()设定表格分组变量以及定义reference group;接着就是各个分析的模块,在analyze()中分别调用上述的自定义函数即可,其中12/36/60月需要多次调用,因此用for循环来实现。最后用build_table()函数调用已完成的layout和数据集来生成最终的表格。
result <- basic_table(
show_colcounts = TRUE,
title = "Table 14.2.1.1: Summary of Efficacy Evaluated"
) |>
split_cols_by("AFB", ref_group = "AFB=Yes") |>
analyze("AFB", a_count_subjd, show_labels = "hidden") |>
analyze("AFB", a_surv_time_func,
var_labels = "Time to event (months)", show_labels = "visible",
extra_args = list(med_tb = surv_med, quant_tb = surv_quant),
table_names = "kmtable"
) |>
analyze("AFB", a_surv_pval_func,
var_labels = "Unstratified log-rank test", show_labels = "visible",
extra_args = list(pval_tb = surv_pval),
table_names = "logrank"
)
time_point <- c(12, 36, 60)
for (i in seq_along(time_point)) {
result <- result |>
analyze("AFB", a_surv_rate_func,
var_labels = paste(time_point[i], "months"), show_labels = "visible",
extra_args = list(rate_tb = surv_rate[[i]], rate_diff_tb = surv_rate_diff[[i]]),
table_names = paste0("timepoint_", time_point[i])
)
}
result |>
build_table(dat %>% mutate(AFB = str_c("AFB=", AFB)))以上是我对于rtables包的粗略理解,详细的教程可参考:https://insightsengineering.github.io/rtables/latest-tag/中的一些文档,以及一些已做分享的presentations(https://insightsengineering.github.io/rtables/latest-tag/#presentations)
现在网上关于用R语言来完成临床分析和图表生成的中文教程相对较少,希望这个简单的分享的能帮助到大家,若有出错的地方还请随时告知
Reference
https://insightsengineering.github.io/rtables/latest-tag/
https://insightsengineering.github.io/tlg-catalog/stable/tables/efficacy/ttet01.html
https://pharmaverse.r-universe.dev/articles/rtables/introduction.html
https://www.pharmasug.org/proceedings/japan2023/PharmaSUG-Japan-2023-05.pdf https://www.r-consortium.org/all-projects/tables-in-clinical-trials-with-r#rtables