The Best Overall Response (BOR) is a very common evaluation of efficacy in oncology trials. Usually, it is defined as the best response among all time-point responses from the treatment start until the first disease progression, in the order of CR, PR, SD, PD, and NE per RECIST 1.1. For non-randomized trials, BOR is not only the best among all responses but also requires confirmation for CR and PR to ensure the result is not a measurement error. More details can be found in the RECIST 1.1 document, which I will not expand on here.
Although there are lots of blogs on Google that will tell you how to derive BOR in SAS, only a few people will use R to do so. This article is to talk about how to implement BOR with or without confirmation in R.
Best Overall Response without confirmation
Firstly, let's look at the programming logic for BOR without confirmation.
- Set to complete response (CR) if one CR exists.
- Set to partial response (PR) if one PR exists .
- Set to stable disease (SD) if one SD exists, which meets the minimum requirement for SD duration from treatment (or randomization) start to the date of the response.
- Set to progressive disease (PD) if one PD exists.
- Set to not estimable (NE) if only NE exists or the response cannot meet minimum SD duration criteria.
Afterwards, you can select the best response above for each subject as the BOR.
Best Overall Response with confirmation
Below are the rules to evaluate the best response where the confirmation of CR and PR is required for BOR deviation, from RECIST guideline (https://ctep.cancer.gov/protocolDevelopment/docs/recist_guideline.pdf)
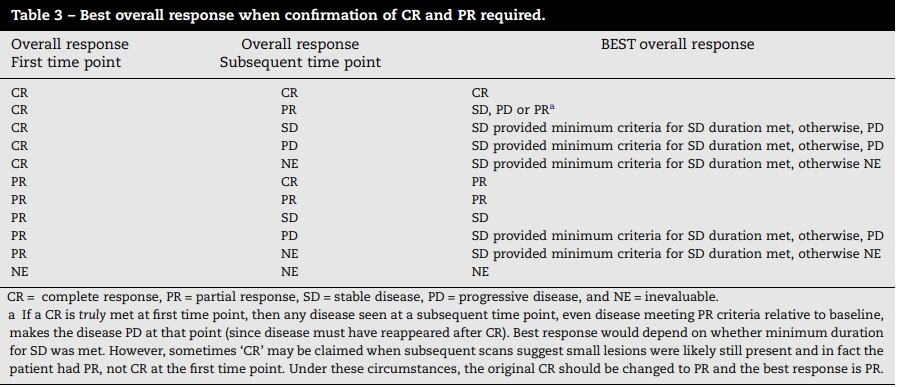
Actually, we don't consider the scenario where the CR is followed by PR, but we must consider the scenario where subsequent response is not sequential. And we also need to consider how many NEs are acceptable between response and confirmatory response.
Thus the programming logic for confirmed BOR can be summarized as following:
- Set to complete response (CR) if there is one confirmatory CR at least a minimum number of days (e.g., 28 days) later, all responses between the two should only be "CR" or "NE", and there are no more than a maximum NE (e.g., one NE) between two responses.
- Set to partial response (PR) if there is one confirmatory CR or PR at least a minimum number of days (e.g., 28 days) later, all responses between the two should only be are "CR", "PR" or "NE", and there are no more than a maximum NE (e.g., one NE) between two PR/CR responses.
- Set to stable disease (SD) if there is one CR, PR or SD that meets the minimum requirement for the duration from treatment (or randomization) start to the date of that response.
- Set to progressive disease (PD) if one PD exists.
- Set to not estimable (NE) if there is at least one CR, PR, SD, NE.
And then like the unconfirmed BOR, you can select the best response above for each subject as the confirmed BOR.
How to implement it in R
I have created a function in stabiot R package following the rules we discussed above. For example, let's try it using the derive_bor() function as shown below. More detials can be found in ?derive_bor.
# This example is referred from `admiral::event_joined`.
adrs <- tibble::tribble(
~USUBJID, ~TRTSDTC, ~ADTC, ~AVALC,
"1", "2020-01-01", "2020-01-01", "PR",
"1", "2020-01-01", "2020-02-01", "CR",
"1", "2020-01-01", "2020-02-16", "NE",
"1", "2020-01-01", "2020-03-01", "CR",
"1", "2020-01-01", "2020-04-01", "SD",
"2", "2019-12-12", "2020-01-01", "SD",
"2", "2019-12-12", "2020-02-01", "PR",
"2", "2019-12-12", "2020-03-01", "SD",
"2", "2019-12-12", "2020-03-13", "CR",
"4", "2019-12-30", "2020-01-01", "PR",
"4", "2019-12-30", "2020-03-01", "NE",
"4", "2019-12-30", "2020-04-01", "NE",
"4", "2019-12-30", "2020-05-01", "PR",
"5", "2020-01-01", "2020-01-01", "PR",
"5", "2020-01-01", "2020-01-10", "PR",
"5", "2020-01-01", "2020-01-20", "PR",
"6", "2020-02-02", "2020-02-06", "PR",
"6", "2020-02-02", "2020-02-16", "CR",
"6", "2020-02-02", "2020-03-30", "PR",
"7", "2020-02-02", "2020-02-06", "PR",
"7", "2020-02-02", "2020-02-16", "CR",
"7", "2020-02-02", "2020-04-01", "NE",
"8", "2020-02-01", "2020-02-16", "PD"
) %>%
dplyr::mutate(
ADT = lubridate::ymd(ADTC),
TRTSDT = lubridate::ymd(TRTSDTC),
PARAMCD = "OVR",
PARAM = "Overall Response by Investigator"
) %>%
dplyr::select(-TRTSDTC)Suppose that we want to calculate the BOR without confirmation and the SD duration is set to 4 weeks, only we simply need to specify ref_start_window = 28.
derive_bor(data = adrs, ref_start_window = 28)
## # A tibble: 7 × 8
## USUBJID ADTC AVALC ADT TRTSDT PARAMCD PARAM AVAL
## <chr> <chr> <chr> <date> <date> <chr> <chr> <dbl>
## 1 1 2020-02-01 CR 2020-02-01 2020-01-01 BOR Best Overall Response 1
## 2 2 2020-03-13 CR 2020-03-13 2019-12-12 BOR Best Overall Response 1
## 3 4 2020-01-01 PR 2020-01-01 2019-12-30 BOR Best Overall Response 2
## 4 5 2020-01-01 PR 2020-01-01 2020-01-01 BOR Best Overall Response 2
## 5 6 2020-02-16 CR 2020-02-16 2020-02-02 BOR Best Overall Response 1
## 6 7 2020-02-16 CR 2020-02-16 2020-02-02 BOR Best Overall Response 1
## 7 8 2020-02-16 PD 2020-02-16 2020-02-01 BOR Best Overall Response 4Suppose that we want to calculate the BOR with confirmation and the SD duration is set to 4 weeks, and the interval of two responses is set to 28 days, we simply need to add ref_interval = 28 and confirm = TRUE
derive_bor(data = adrs, ref_start_window = 28, ref_interval = 28, confirm = TRUE)
## # A tibble: 7 × 8
## USUBJID ADTC AVALC ADT TRTSDT PARAMCD PARAM AVAL
## <chr> <chr> <chr> <date> <date> <chr> <chr> <dbl>
## 1 1 2020-02-01 CR 2020-02-01 2020-01-01 CBOR Confirmed Best Overall Response 1
## 2 2 2020-02-01 SD 2020-02-01 2019-12-12 CBOR Confirmed Best Overall Response 3
## 3 4 2020-05-01 SD 2020-05-01 2019-12-30 CBOR Confirmed Best Overall Response 3
## 4 5 2020-01-01 NE 2020-01-01 2020-01-01 CBOR Confirmed Best Overall Response 5
## 5 6 2020-02-06 PR 2020-02-06 2020-02-02 CBOR Confirmed Best Overall Response 2
## 6 7 2020-02-06 NE 2020-02-06 2020-02-02 CBOR Confirmed Best Overall Response 5
## 7 8 2020-02-16 PD 2020-02-16 2020-02-01 CBOR Confirmed Best Overall Response 4If we don't want any NE between the response and confirmatory response in addition to the above conditions, we can simply add max_ne = 0.
derive_bor(data = adrs, ref_start_window = 28, ref_interval = 28, confirm = TRUE, max_ne = 0)
## # A tibble: 7 × 8
## USUBJID ADTC AVALC ADT TRTSDT PARAMCD PARAM AVAL
## <chr> <chr> <chr> <date> <date> <chr> <chr> <dbl>
## 1 1 2020-01-01 PR 2020-01-01 2020-01-01 CBOR Confirmed Best Overall Response 2
## 2 2 2020-02-01 SD 2020-02-01 2019-12-12 CBOR Confirmed Best Overall Response 3
## 3 4 2020-05-01 SD 2020-05-01 2019-12-30 CBOR Confirmed Best Overall Response 3
## 4 5 2020-01-01 NE 2020-01-01 2020-01-01 CBOR Confirmed Best Overall Response 5
## 5 6 2020-02-06 PR 2020-02-06 2020-02-02 CBOR Confirmed Best Overall Response 2
## 6 7 2020-02-06 NE 2020-02-06 2020-02-02 CBOR Confirmed Best Overall Response 5
## 7 8 2020-02-16 PD 2020-02-16 2020-02-01 CBOR Confirmed Best Overall Response 4The above all are my summries for BOR calculation. If there is any problem or error, please email me to let me know, or leave your issues in the https://github.com/kaigu1990/stabiot/issues.
At the very least, I'd like to appreciate the admiral R package, I have learned more programming skills for BOR calculation from admiral::derive_extreme_event().
Reference
https://github.com/pharmaverse/admiral
https://www.pharmasug.org/proceedings/2023/QT/PharmaSUG-2023-QT-047.pdf
https://www.pharmasug.org/proceedings/2020/DV/PharmaSUG-2020-DV-066.pdf
https://ctep.cancer.gov/protocolDevelopment/docs/recist_guideline.pdf
https://www.lexjansen.com/pharmasug-cn/2021/SR/Pharmasug-China-2021-SR038.pdf